Tutorial 5b – Inhomogeneous one-dimensional log-Normal random field
In this tutorial we use Fesslix to discretize a inhomogeneous one-dimensional log-Normal random field.
For this tutorial you must have a 64-bit version of Python 2.7 installed on your system.
You can download the latest 2.7 release from here: https://www.python.org/downloads/windows/.
Table of Contents
1 Random field properties
The goal is to represent a log-Normal random field that has mean 10 and standard deviation function
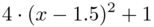 .
We assume that the random field can be transformed to an underlying Gaussian random field; i.e.,
by taking the natural logarithm of the log-Normal random field.
The auto-correlation coefficient function of the underlying Gaussian random field is:
.
We assume that the random field can be transformed to an underlying Gaussian random field; i.e.,
by taking the natural logarithm of the log-Normal random field.
The auto-correlation coefficient function of the underlying Gaussian random field is:
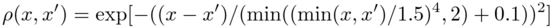 .
The domain of interest has a length of 3m.
.
The domain of interest has a length of 3m.
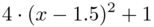 .
We assume that the random field can be transformed to an underlying Gaussian random field; i.e.,
by taking the natural logarithm of the log-Normal random field.
The auto-correlation coefficient function of the underlying Gaussian random field is:
.
We assume that the random field can be transformed to an underlying Gaussian random field; i.e.,
by taking the natural logarithm of the log-Normal random field.
The auto-correlation coefficient function of the underlying Gaussian random field is:
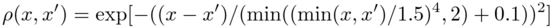 .
The domain of interest has a length of 3m.
.
The domain of interest has a length of 3m.
2 Step by Step Instruction
Note: This tutorial uses the Python plotting library matplotlib. If you do not have Python and matplotlib installed on your system, you have to comment-out the corresponding lines in the Fesslix parameter file.
2.1 Random field discretization
Fesslix parameter file
#! =============================== #! Load the FE module #! =============================== loadlib "fem"; #! =============================== #! Initial definitions #! =============================== const l = 3; # [m] length of random field domain const mu = 10; # mean value of random field var sd = 4*(gx-1.5)^2+1; # std. dev. function of random field # auto-correlation coefficient function of underlying Gaussian field var corr = exp(-1*(deltap/(min((min(gx,gx2)/1.5)^4,2)+0.1))^2); const M = 10; # number of terms in the random field expansion # mean of underlying Gaussian field fun lambda(2) = ln($1)-ln(($2/$1)^2+1)/2; # std. dev. of underlying Gaussian field fun chi(2) = sqrt(ln(($2/$1)^2+1)); #! =============================== #! Define the mesh #! =============================== # The mesh becomes finer from right to left. const Nel = 20; const x = 1; fun xf(2) = $1*(1-1/$2)^(($2/Nel)^8*1+1); for (i=Nel;i>0.5;i-1) { node i [l*x]; funplot i, l*x; const x = xf(x,i); }; node 0 [0]; for (i=1;i<Nel+0.5;i+1) { edge i i-1 i; }; #! =============================== #! Random field discretization (FE-KL) #! =============================== const pO = 10; # polynomial order of the FE shape functions const MO = 0.1; # 1/MO is number of sub-intervals in outer integration loop # type of discretization: finite element KL expansion rf_new 1 kl_fem_gauss mu lambda(mu,sd) sd chi(mu,sd) rhocor CORR; # Assign a geometry to the field: for (i=1;i<Nel+0.5;i+1) { rf_elset 1 edge i porder=PO MAXSIZEO=MO {dolog=false;}; }; # Obtain the random field discretization rf_solve 1 dim=M calcerr=false; funplot #rf_err(1,1), rf_err(1,101), # Var-based measure - used instead of '1' as std.dev. varies rf_err(1,2), # Cov-based measure rf_err(1,3); # error measure 3
2.2 Output Eigenvalues
Fesslix parameter file
#! =============================== #! Output eigenvalues #! =============================== const NEV = M; sfor (i;NEV) { funplot i, rf_eigenvalue(1,i); };
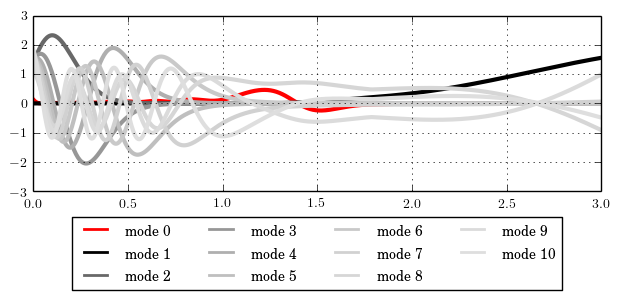
2.3 Plot the eigenmodes of the random fields
Fesslix parameter file
#! =============================== #! Plot the eigenmodes of the random fields #! =============================== #! ------------------------------- #! output the eigenmodes to files #! ------------------------------- const ppd = 500; # number of points to use for plotting mtxconst_new ppd1(3,1,ppd/Nel); # ... for 2nd random field filestream fs ("output_rf2.dat"); rf_plot 1 { ppdim = ppd1; stream=fs; }; #! ------------------------------- #! invoke the tool matplotlib #! ------------------------------- # You can comment this section out if you do not have # 'Python' or 'matplotlib' installed on your system. # load the python-module loadlib "Python"; # add current path to Python search path python_path $pwd(); # load the module 'tutorial_1d_lsf' python_module tutorial_5b_plot; # link the Python function to Fesslix python_fun plot_fun = tutorial_5b_plot.PlotModes; # start the plotting calc plot_fun();
Again, as in the previous tutorial (Tutorial 5a), Python with the plotting library matplotlib is used.
The corresponding Python code is given at the end of this tutorial.
2.4 Plot covariance around 1.5m
Fesslix parameter file
#! =============================== #! Plot covariance around 1.5 #! =============================== # of the underlying Gaussian random field!!! #! ------------------------------- #! output the covariance structure #! ------------------------------- filestream fs ("output_rf_cov.dat"); rf_plotcov 1 [1.5] {stream=fs;}; #! ------------------------------- #! invoke the tool matplotlib #! ------------------------------- # You can comment this section out if you do not have # 'Python' or 'matplotlib' installed on your system. # link the Python function to Fesslix python_fun plot_cov = tutorial_5b_plot.PlotCov; calc plot_cov();
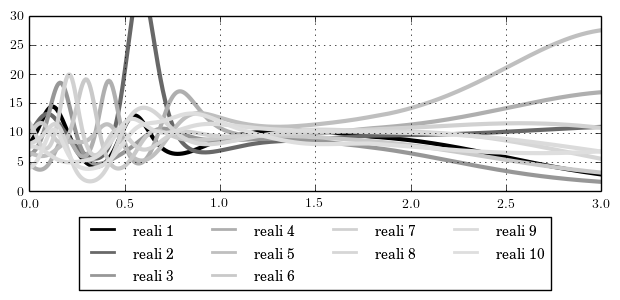
2.5 Plot realizations
Fesslix parameter file
#! =============================== #! Plot realizations #! =============================== #! ------------------------------- #! generate realizations #! ------------------------------- const Nrep = 10; const Npel = 20; mtxconst_new resv(Nel*(Npel+1),Nrep+1,0); sfor (j;Nrep) { const c = 0; rnd_smp "rf_1"; for (i=1;i<Nel+0.5;i+1) { for (lx=-1;lx<=1;lx+2/Npel) { const res = geocalc(edge,i,exp(rf(1)),lx); mtxcoeff resv(c,0) = gx; mtxcoeff resv(c,j) = res; const c += 1; }; }; }; #! ------------------------------- #! output the realizations #! ------------------------------- filestream fs("output_reali.dat"); sfor (j;Nel*(Npel+1)) { mtxconst_sub vsub = resv(row:j); funplot {vsub} { stream=fs; }; }; #! ------------------------------- #! invoke the tool matplotlib #! ------------------------------- # You can comment this section out if you do not have # 'Python' or 'matplotlib' installed on your system. # link the Python function to Fesslix python_fun plot_reali = tutorial_5b_plot.PlotReali; calc plot_reali();
2.6 Python code
The Python code used to generate the plots is:
Python code "tutorial_5b_plot.py"
# -*- coding: utf-8 -*- import flx; import matplotlib.pyplot as plt import matplotlib as mpl import numpy as np import matplotlib.patches as mpatches from matplotlib.backends.backend_pgf import FigureCanvasPgf mpl.backend_bases.register_backend('pdf', FigureCanvasPgf) pgf_with_pdflatex = { "pgf.texsystem": "pdflatex", "pgf.preamble": [ r"\usepackage[utf8x]{inputenc}", r"\usepackage[T1]{fontenc}", # r"\usepackage{cmbright}", ] } mpl.rcParams.update(pgf_with_pdflatex) def PlotModes(): # ===================================== # Read data from files # ===================================== dfile = r'output_rf2.dat' m = {} x, er, m[1], m[2], m[3], m[4], m[5], m[6], m[7], m[8], m[9], m[10] = np.genfromtxt( dfile, unpack=True, usecols=(0,3,4,5,6,7,8,9,10,11,12,13)) # ===================================== # Do the plotting # ===================================== plt.rc('text', usetex=True) plt.rc('font', family='roman', size=10) plt.figure(1, figsize=(6.3,2.5)) plt.grid(True) plt.plot(x, er, linewidth=3, color='red', label=r'mode 0') for i in range (1,11): plt.plot(x, m[i], linewidth=3, color="{:.9f}".format(pow(1.-(1./i),1.3)), label=r'mode '+"{:.0f}".format(i)) plt.xlabel(r'$x$') art = [] legend = plt.legend(loc=9, bbox_to_anchor=(0.5, -0.1),ncol=4) art.append(legend) for label in legend.get_texts(): label.set_fontsize('medium') for label in legend.get_lines(): label.set_linewidth(2) # the legend line width plt.tight_layout() plt.savefig('tutorial_5b_plot_modes.pdf', format='pdf', additional_artists=art, bbox_inches="tight") plt.savefig('tutorial_5b_plot_modes.png', format='png', additional_artists=art, bbox_inches="tight") plt.show() return 0; def PlotReali(): # ===================================== # Read data from files # ===================================== dfile = r'output_reali.dat' m = {} x, m[1], m[2], m[3], m[4], m[5], m[6], m[7], m[8], m[9], m[10] = np.genfromtxt( dfile, unpack=True, usecols=(0,1,2,3,4,5,6,7,8,9,10)) # ===================================== # Do the plotting # ===================================== plt.rc('text', usetex=True) plt.rc('font', family='roman', size=10) plt.figure(1, figsize=(6.3,2.5)) plt.grid(True) for i in range (1,11): plt.plot(x, m[i], linewidth=3, color="{:.9f}".format(pow(1.-(1./i),1.3)), label=r'reali '+"{:.0f}".format(i)) plt.xlabel(r'$x$') plt.ylim([0,30]) art = [] legend = plt.legend(loc=9, bbox_to_anchor=(0.5, -0.1),ncol=4) art.append(legend) for label in legend.get_texts(): label.set_fontsize('medium') for label in legend.get_lines(): label.set_linewidth(2) # the legend line width plt.tight_layout() plt.savefig('tutorial_5b_plot_reali.pdf', format='pdf', additional_artists=art, bbox_inches="tight") plt.savefig('tutorial_5b_plot_reali.png', format='png', additional_artists=art, bbox_inches="tight") plt.show() return 0; def PlotCov(): # ===================================== # Read data from files # ===================================== dfile = r'output_rf_cov.dat' m = {} x, m[1], m[2], m[3] = np.genfromtxt(dfile, unpack=True, usecols=(0,3,4,5)) # ===================================== # Do the plotting # ===================================== plt.rc('text', usetex=True) plt.rc('font', family='roman', size=10) plt.figure(1, figsize=(6.3,2.5)) plt.grid(True) plt.plot(x, m[1], linewidth=3, color="red", label=r'cov approx.') plt.plot(x, m[2], linewidth=3, color="black", label=r'cov analytical') plt.plot(x, m[3], linewidth=3, color="orange", label=r'difference') plt.xlabel(r'$x$') art = [] legend = plt.legend(loc=9, bbox_to_anchor=(0.5, -0.1),ncol=4) art.append(legend) for label in legend.get_texts(): label.set_fontsize('medium') for label in legend.get_lines(): label.set_linewidth(2) # the legend line width plt.tight_layout() plt.savefig('tutorial_5b_plot_cov.pdf', format='pdf', additional_artists=art, bbox_inches="tight") plt.savefig('tutorial_5b_plot_cov.png', format='png', additional_artists=art, bbox_inches="tight") plt.show() return 0;